G protein-coupled receptor (GPCR), also known as the seven transmembrane receptor, is the largest receptor family in the human genome. There are more than 800 genes encoding GPCR, which are responsible for about 80% of transmembrane signal transduction, and participate in the regulation of many pathological and physiological processes in the human body. At present, more than 30% of prescription drugs are targeting GPCR. GPCR transforms extracellular stimulations into intracellular signals mainly through G proteins and arrestins. In these processes, G proteins were able to regulate the level of intracellular second messenger, whereas arrestins could recruit different downstream signaling molecules to desensitize the receptor, facilitate endocytosis, and other G protein independent signal transduction pathways. However, the precise regulation mechanism of GPCR by arrestin is still unclear, which makes it difficult to develop and apply the biased ligands of GPCR to treat human diseases.
In 2014, research group led by Professor Robert J. Lefkowitz at Duke University found that arrstin mainly binds to GPCR in two ways: through the "hanging configuration" which binds solely to the phosphorylated C-terminal tail of the receptor, or the "snuggly configuration" by forming extensive interactions with the seven transmembrane core of the receptor. However, in the "snuggly configuration", how different ligands of a single GPCR modulate different functional selective conformations of arrestin is still unclear.
After four years of hard work, Professor Sun Jinpeng and Professor Wang Jiangyun at the Chinese Academy of Sciences, published a research paper entitled“DeSiphering receptor core-induced and ligand-dependent conformational changes in arrestin via genetic encoded trimethylsilyl1H-NMR probe” in Nature Communications.This study provide direct evidence that ligands of a single receptor could selectively regulate different functional relative configurations of arrestin directly through the interactions of seven transmembrane helix core (TM core) of the GPCR with the arrestin at residue resolution.
Since a large amount of proteins are needed to monitor the configurations of receptor/arrestin complexes, new methods must be developed to reduce the membrane protein usage. Therefore, our research group has developed a new one-dimensional hydrogen NMR spectrum probe, which can obtain the conformational information of GPCRs signal complex in a short time with low protein concentration. The common elements in natural proteins are nitrogen, oxygen, phosphorus, sulfur, and carbon, but not silicon, the element often used in computer. Through the gene codon expansion technology, the team developed an innovative technology to introduce 4-methylphenylpropanosilicone into protein, named “Desipher”, in which the letter c in decipher was replaced by letter s in accordance with silicon element to highlight the innovation of the method. This labeling method combined with the use of 950 MHz nuclear magnetic resonance spectrometer, can obtain the dynamic conformation changes of arrestin protein at the residue level in low protein concentration (as low as 5 μ M) and in a short time (approximately 10 minutes). This labeling method provides a powerful tool for the study of GPCR complex.
With this innovative probe, the team demonstrated for the first time at residue resolution, that different ligands are capable of modulate different functional related arrestin conformations through the seven transmembrane core (TM core) of GPCR . This study shows that the regulation of arrestin by ligands can circumvent the receptor phosphorylation barcode, which has also extensively studied by the same group in previous studies, thus provides new ideas for the development of biased drugs targeting to arrestin mediated GPCR functions.
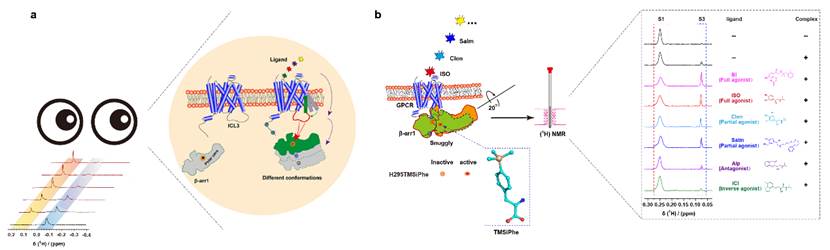
Desiphering the ligands interact with the transmembrane core (TM core) of GPCR and directly regulate the conformational changes of arrestin
Liu Qi, He Qingtao and Yang Fan at School of Basic Medical Sciences of Shandong University, Lv Xiaoxuan in Institute of Biophysics and Zhu Zhongliang at School of Life Sciences of University of Science and Technology of China are co-first authors in this paper; Professor Sun Jinpeng and Professor Wang Jiangyun are co-corresponding authors.
This research has been funded and supported by the National Science Foundation for Distinguished Young Scholars and the National Natural Science Foundation of China.
Link to original text:https://www.nature.com/articles/s41467-020-18433-5
Written by: He Qingtao
Edited by: Che Huiqing